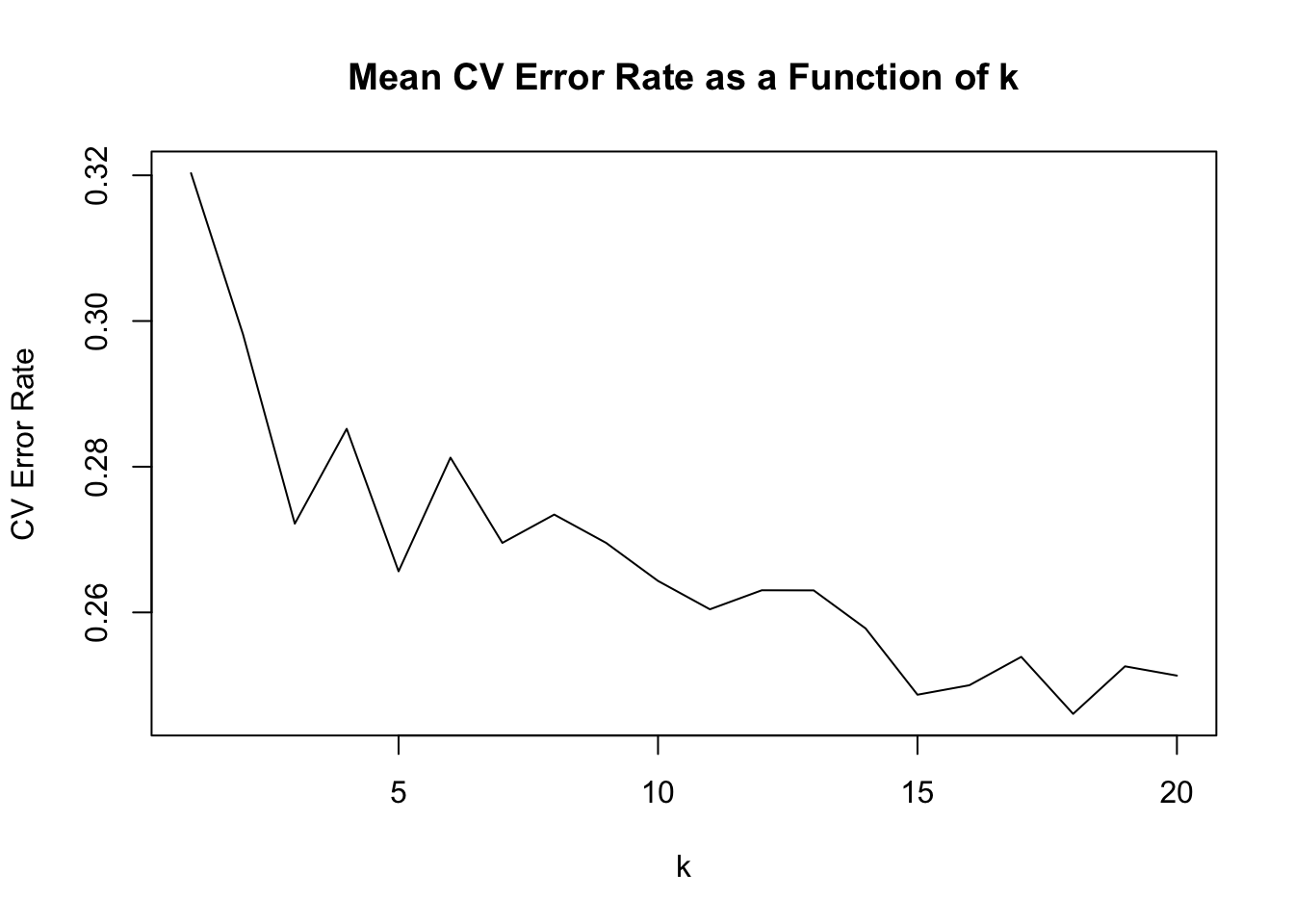
[,1] [,2] [,3] [,4] [,5]
[1,] 0.3636364 0.3246753 0.2857143 0.3137255 0.3137255
[2,] 0.2857143 0.3051948 0.2727273 0.3137255 0.3137255
[3,] 0.2727273 0.2727273 0.2337662 0.2745098 0.3071895
[4,] 0.2597403 0.2727273 0.2857143 0.2614379 0.3464052
[5,] 0.2402597 0.2922078 0.2532468 0.2483660 0.2941176
[6,] 0.2662338 0.3116883 0.2662338 0.2549020 0.3071895
[7,] 0.2207792 0.2987013 0.2792208 0.2549020 0.2941176
[8,] 0.2467532 0.2922078 0.2857143 0.2549020 0.2875817
[9,] 0.2207792 0.2987013 0.2792208 0.2614379 0.2875817
[10,] 0.2402597 0.2922078 0.2597403 0.2745098 0.2549020
[11,] 0.2272727 0.2857143 0.2597403 0.2549020 0.2745098
[12,] 0.2207792 0.2857143 0.2662338 0.2549020 0.2875817
[13,] 0.2402597 0.2727273 0.2727273 0.2549020 0.2745098
[14,] 0.2337662 0.2727273 0.2727273 0.2352941 0.2745098
[15,] 0.2337662 0.2597403 0.2467532 0.2418301 0.2614379
[16,] 0.2662338 0.2467532 0.2337662 0.2483660 0.2549020
[17,] 0.2597403 0.2597403 0.2467532 0.2418301 0.2614379
[18,] 0.2597403 0.2272727 0.2662338 0.2222222 0.2549020
[19,] 0.2662338 0.2402597 0.2532468 0.2222222 0.2810458
[20,] 0.2597403 0.2337662 0.2402597 0.2483660 0.2745098
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 0.2857143 0.2727273 0.3246753 0.2727273 0.3246753 0.2597403 0.3636364
[2,] 0.3506494 0.2467532 0.2597403 0.2987013 0.2597403 0.2857143 0.4675325
[3,] 0.2207792 0.2337662 0.3246753 0.2337662 0.3376623 0.1948052 0.3506494
[4,] 0.2597403 0.2467532 0.2597403 0.2857143 0.2987013 0.1558442 0.3506494
[5,] 0.2727273 0.2857143 0.3246753 0.2467532 0.3506494 0.1688312 0.3246753
[6,] 0.2857143 0.2597403 0.3116883 0.2467532 0.3376623 0.1818182 0.2987013
[7,] 0.2727273 0.2337662 0.3246753 0.2337662 0.2857143 0.1688312 0.2857143
[8,] 0.3116883 0.2077922 0.2857143 0.2467532 0.3116883 0.1818182 0.3116883
[9,] 0.2467532 0.1818182 0.2987013 0.2467532 0.2987013 0.1818182 0.2857143
[10,] 0.2337662 0.1818182 0.2987013 0.2337662 0.2727273 0.2207792 0.2727273
[11,] 0.2467532 0.2077922 0.3246753 0.2077922 0.3116883 0.1948052 0.2987013
[12,] 0.2467532 0.1818182 0.2857143 0.2337662 0.3246753 0.1818182 0.2467532
[13,] 0.2207792 0.1688312 0.2987013 0.2727273 0.2987013 0.2077922 0.2597403
[14,] 0.2467532 0.1948052 0.2987013 0.2467532 0.2987013 0.2337662 0.2597403
[15,] 0.2467532 0.1818182 0.2727273 0.2207792 0.2987013 0.2337662 0.2727273
[16,] 0.2077922 0.1818182 0.2857143 0.2337662 0.3116883 0.2467532 0.2987013
[17,] 0.2207792 0.1818182 0.2727273 0.2337662 0.3376623 0.2337662 0.2597403
[18,] 0.2467532 0.1818182 0.2727273 0.2597403 0.3246753 0.2337662 0.2597403
[19,] 0.2207792 0.1818182 0.2727273 0.2207792 0.3246753 0.2467532 0.2597403
[20,] 0.2077922 0.1818182 0.2857143 0.2467532 0.3116883 0.2077922 0.2857143
[21,] 0.2337662 0.1948052 0.2857143 0.2077922 0.3246753 0.2207792 0.2597403
[22,] 0.2077922 0.1818182 0.3376623 0.2337662 0.3376623 0.2207792 0.2727273
[23,] 0.1948052 0.1818182 0.3116883 0.2337662 0.3116883 0.2077922 0.2727273
[24,] 0.2207792 0.1428571 0.3116883 0.2077922 0.3246753 0.2077922 0.2337662
[25,] 0.2207792 0.1948052 0.3246753 0.2077922 0.3246753 0.2077922 0.2467532
[26,] 0.2207792 0.1948052 0.3376623 0.2337662 0.2727273 0.1818182 0.2727273
[27,] 0.2467532 0.1818182 0.3376623 0.2337662 0.2727273 0.1948052 0.2857143
[28,] 0.2337662 0.1818182 0.3246753 0.2207792 0.2857143 0.1948052 0.2857143
[29,] 0.2467532 0.1948052 0.3506494 0.2467532 0.2987013 0.2077922 0.2467532
[30,] 0.2337662 0.1948052 0.3506494 0.2207792 0.3116883 0.2077922 0.2727273
[31,] 0.2207792 0.1948052 0.3376623 0.2337662 0.2987013 0.2207792 0.2727273
[32,] 0.2207792 0.1818182 0.3376623 0.2207792 0.2987013 0.2207792 0.2597403
[33,] 0.1948052 0.1818182 0.3376623 0.2467532 0.2987013 0.2077922 0.2597403
[34,] 0.1818182 0.1948052 0.3506494 0.2077922 0.2857143 0.1948052 0.2597403
[35,] 0.1818182 0.1818182 0.3376623 0.2207792 0.2987013 0.2077922 0.2337662
[36,] 0.2077922 0.1818182 0.3376623 0.1948052 0.3116883 0.1948052 0.2337662
[37,] 0.1948052 0.1818182 0.3506494 0.2337662 0.2987013 0.2207792 0.2337662
[38,] 0.2077922 0.1818182 0.3246753 0.2467532 0.2987013 0.2337662 0.2337662
[39,] 0.2207792 0.1948052 0.3246753 0.2077922 0.2987013 0.2337662 0.2597403
[40,] 0.2077922 0.1948052 0.3116883 0.2207792 0.2987013 0.2337662 0.2597403
[41,] 0.2077922 0.1948052 0.3246753 0.2077922 0.2987013 0.2337662 0.2597403
[42,] 0.2207792 0.1948052 0.3376623 0.1818182 0.3116883 0.2207792 0.2597403
[43,] 0.2207792 0.1818182 0.3246753 0.1948052 0.3116883 0.2337662 0.2467532
[44,] 0.1948052 0.1948052 0.3116883 0.2077922 0.3116883 0.2597403 0.2467532
[45,] 0.2337662 0.1818182 0.3116883 0.1948052 0.2987013 0.2207792 0.2467532
[46,] 0.2337662 0.1948052 0.2987013 0.1948052 0.2987013 0.2337662 0.2597403
[47,] 0.2467532 0.1948052 0.3376623 0.2207792 0.2987013 0.2467532 0.2467532
[48,] 0.2467532 0.1688312 0.3376623 0.2077922 0.2987013 0.2207792 0.2467532
[49,] 0.2337662 0.1688312 0.3376623 0.2077922 0.2857143 0.2467532 0.2467532
[50,] 0.2337662 0.1688312 0.3376623 0.2207792 0.2857143 0.2337662 0.2337662
[,8] [,9] [,10]
[1,] 0.2727273 0.3026316 0.2631579
[2,] 0.2467532 0.3289474 0.3157895
[3,] 0.3246753 0.2105263 0.2500000
[4,] 0.2207792 0.3026316 0.2368421
[5,] 0.2337662 0.2894737 0.2368421
[6,] 0.2597403 0.3157895 0.2368421
[7,] 0.2597403 0.2763158 0.2763158
[8,] 0.2597403 0.3157895 0.2105263
[9,] 0.2207792 0.2631579 0.2368421
[10,] 0.2077922 0.3026316 0.2500000
[11,] 0.1948052 0.2631579 0.2894737
[12,] 0.1948052 0.2894737 0.2763158
[13,] 0.2077922 0.2631579 0.2368421
[14,] 0.2077922 0.2763158 0.2763158
[15,] 0.1818182 0.2500000 0.2631579
[16,] 0.2207792 0.2105263 0.2500000
[17,] 0.2207792 0.2236842 0.2631579
[18,] 0.2337662 0.2631579 0.2894737
[19,] 0.1948052 0.2368421 0.2763158
[20,] 0.2077922 0.2368421 0.2631579
[21,] 0.2077922 0.2500000 0.2631579
[22,] 0.2207792 0.2500000 0.2631579
[23,] 0.2077922 0.2500000 0.2631579
[24,] 0.2077922 0.2631579 0.2894737
[25,] 0.2077922 0.2500000 0.2763158
[26,] 0.2207792 0.2631579 0.2894737
[27,] 0.1948052 0.2631579 0.2763158
[28,] 0.2207792 0.2368421 0.2894737
[29,] 0.1948052 0.2500000 0.3026316
[30,] 0.1948052 0.2500000 0.2894737
[31,] 0.1818182 0.2631579 0.3026316
[32,] 0.2077922 0.2500000 0.3157895
[33,] 0.1818182 0.2500000 0.2894737
[34,] 0.2077922 0.2631579 0.2894737
[35,] 0.2207792 0.2500000 0.2894737
[36,] 0.2207792 0.2368421 0.2763158
[37,] 0.2207792 0.2500000 0.2763158
[38,] 0.2077922 0.2368421 0.2894737
[39,] 0.2207792 0.2500000 0.2763158
[40,] 0.2207792 0.2631579 0.3157895
[41,] 0.2077922 0.2500000 0.2763158
[42,] 0.2207792 0.2500000 0.2894737
[43,] 0.2077922 0.2500000 0.2894737
[44,] 0.2207792 0.2631579 0.3026316
[45,] 0.2077922 0.2631579 0.2894737
[46,] 0.1948052 0.2631579 0.2894737
[47,] 0.2077922 0.2763158 0.2894737
[48,] 0.2077922 0.2631579 0.3026316
[49,] 0.2077922 0.2631579 0.3026316
[50,] 0.2077922 0.2631579 0.3026316